Research areas/focus
DrugDiff

Our research group has been developing DrugDiff, a latent diffusion model for the generation of small drug-like molecules inspired by advancements in image generation. By including guidance into the generation process, users can effectively influence specific molecular properties in the produced compounds. For instance, our experiments have demonstrated responsive control over properties such as logP, synthetic accessibility, and quantitative estimation of drug-likeness. By modulating the 'guidance strength', we observed discernible shifts in the property distributions of generated molecules and our results show good generalization across chemically diverse small molecule datasets. We are currently exploring the full potential of DrugDiff's adaptable architecture, including its modularity and property agnosticism, to further enhance controlled molecular generation.
PriSyn
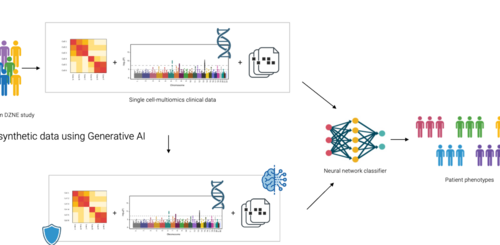
The PriSyn project is a collaborative initiative focused on transforming the utilization of health data for research purposes. While acknowledging the importance of medical data such as genetic information and blood counts, the strict limitations and controlled access of this data brings forward the challenge of data sharing and usability across different sources. The key collaborators in this project are the German Center for Neurodegenerative Diseases (DZNE), CISPA Helmholtz Center for Information Security, startup QuantPi, and IT company Hewlett Packard Enterprise (HPE). Their main focus will be to address the efficient and privacy-preserving medical data utilization while maintaining privacy protection. The project thereby proposes an innovative method of anonymizing data, which unlike previous methods that involved noise addition to the data, uses machine learning models that are trained under the mechanism of differential privacy to produce artificial data. The synthetic data generated will reflect the statistical properties of the real data while ensuring privacy is adhered to and a wider accessibility of the data.
CISPA will play the key role in generating the artificial data. DZNE will focus on building a single-cell multi-center dataset which will incorporate clinical metadata, single-cell transcriptome data and genotypes. Additionally, they will be involved in creating a neurodegenerative classifier, and as part of preprocessing, create biologically plausible metrics to evaluate the models created by the synthetic data. QuantPi will also play a crucial role in evaluating the quality of synthetic data for various use cases. This will involve identifying measures and benchmark experiments that can accurately analyze the trade-off between privacy protection and data utility. HPE will be responsible for ensuring that the generative models developed within the project are efficiently integrated into hardware solutions that are user-friendly and secure.
Small Molecule Autoencoders
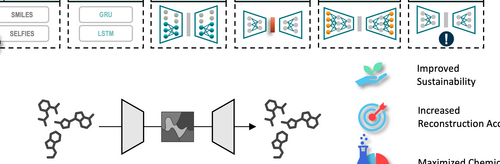
Autoencoders are commonly employed to create embeddings for molecular data used in subsequent deep learning applications. However, there is a notable gap in assessing the quality of chemical information captured in the latent spaces, and the selection of model architectures often appears to be random. Such unoptimized architectures can compromise the integrity of the latent space and lead to excessive energy usage during training, rendering the models less sustainable. We have carried out a series of systematic experiments to investigate the impact of autoencoder architecture on both the accuracy of molecular reconstruction and the quality of the latent space. Our findings indicate that by optimizing the architecture, we can achieve parity in performance with general architectures while utilizing 97% less data and cutting energy consumption by approximately 36%. Furthermore, our experiments revealed that using SELFIES for molecular representation led to a decrease in reconstruction accuracy compared to SMILES representations. Conversely, training with enumerated SMILES significantly enhanced the quality of the latent space.
PRO-GENE-GEN
Genetic data is highly privacy sensitive information and therefore is protected under stringent legal regulations, making sharing it burdensome. However, leveraging genetic information bears great potential in diagnosis and treatment of diseases and is essential for personalized medicine to become a reality. While privacy preserving mechanisms have been introduced, they either pose significant overheads or fail to fully protect the privacy of sensitive patient data. This reduces the ability to share data with the research community which hinders scientific discovery as well as reproducibility of results. Hence, we propose a different approach using synthetic data sets that share the properties of patient data sets while respecting the privacy. We achieve this by leveraging the latest advances in generative modeling to synthesize virtual cohorts. Such synthetic data can be analyzed with established tool chains, repeated access does not affect the privacy budget and can even be shared openly with the research community. While generative modeling of high dimensional data like genetic data has been prohibitive, latest developments in deep generative models have shown a series of success stories on a wide range of domains. The project's finding are summarized in our paper, which was accepted at PoPETS 2024.
Swarm Learning
We are part of the Swarm Learning activities at DZNE and work on several projects related to high-dimensional flow cytometry on a global scale, medical imaging, and joint fine-tuning of large language models. Furthermore, we investigate the use of swarm learning in generative models.
Modular HPC
We use HPC and GPU clusters at DZNE, develop tools and pipelines and engage with the DZNE community on transferring computing tasks to our clusters using containerization following best practices in reproducibility and research software engineering. Based on our strong computational background, we continuously evaluate new technologies like FPGAs (Field Programmable Gate Arrays), which can take energy-efficiency in machine learning and other tasks to the next level, and work on their integration into DZNE research projects.